
A | B | C | D | E | F | G | H | CH | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9
| Part of a series on |
| Genetics |
|---|
 |

In the fields of molecular biology and genetics, a genome is all the genetic information of an organism.[1] It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as regulatory sequences (see non-coding DNA), and often a substantial fraction of junk DNA with no evident function.[2][3] Almost all eukaryotes have mitochondria and a small mitochondrial genome.[2] Algae and plants also contain chloroplasts with a chloroplast genome.
The study of the genome is called genomics. The genomes of many organisms have been sequenced and various regions have been annotated. The Human Genome Project was started in October 1990, and then reported the sequence of the human genome in April 2003,[4] although the initial "finished" sequence was missing 8% of the genome consisting mostly of repetitive sequences.[5]
With advancements in technology that could handle sequencing of the many repetitive sequences found in human DNA that were not fully uncovered by the original Human Genome Project study, scientists reported the first end-to-end human genome sequence in March 2022.[6]
Origin of the term
The term genome was created in 1920 by Hans Winkler,[7] professor of botany at the University of Hamburg, Germany. The website Oxford Dictionaries and the Online Etymology Dictionary suggest the name is a blend of the words gene and chromosome.[8][9][10][11] However, see omics for a more thorough discussion. A few related -ome words already existed, such as biome and rhizome, forming a vocabulary into which genome fits systematically.[12]
Definition
It's very difficult to come up with a precise definition of "genome." It usually refers to the DNA (or sometimes RNA) molecules that carry the genetic information in an organism but sometimes it is difficult to decide which molecules to include in the definition; for example, bacteria usually have one or two large DNA molecules (chromosomes) that contain all of the essential genetic material but they also contain smaller extrachromosomal plasmid molecules that carry important genetic information. The definition of 'genome' that's commonly used in the scientific literature is usually restricted to the large chromosomal DNA molecules in bacteria.[13]
Nuclear genome
Eukaryotic genomes are even more difficult to define because almost all eukaryotic species contain nuclear chromosomes plus extra DNA molecules in the mitochondria. In addition, algae and plants have chloroplast DNA. Most textbooks make a distinction between the nuclear genome and the organelle (mitochondria and chloroplast) genomes so when they speak of, say, the human genome, they are only referring to the genetic material in the nucleus.[2][14] This is the most common use of 'genome' in the scientific literature.
Ploidy
Most eukaryotes are diploid, meaning that there are two of each chromosome in the nucleus but the 'genome' refers to only one copy of each chromosome. Some eukaryotes have distinctive sex chromosomes, such as the X and Y chromosomes of mammals, so the technical definition of the genome must include both copies of the sex chromosomes. For example, the standard reference genome of humans consists of one copy of each of the 22 autosomes plus one X chromosome and one Y chromosome.[15]
Sequencing and mapping
A genome sequence is the complete list of the nucleotides (A, C, G, and T for DNA genomes) that make up all the chromosomes of an individual or a species. Within a species, the vast majority of nucleotides are identical between individuals, but sequencing multiple individuals is necessary to understand the genetic diversity.

In 1976, Walter Fiers at the University of Ghent (Belgium) was the first to establish the complete nucleotide sequence of a viral RNA-genome (Bacteriophage MS2). The next year, Fred Sanger completed the first DNA-genome sequence: Phage Φ-X174, of 5386 base pairs.[16] The first bacterial genome to be sequenced was that of Haemophilus influenzae, completed by a team at The Institute for Genomic Research in 1995. A few months later, the first eukaryotic genome was completed, with sequences of the 16 chromosomes of budding yeast Saccharomyces cerevisiae published as the result of a European-led effort begun in the mid-1980s. The first genome sequence for an archaeon, Methanococcus jannaschii, was completed in 1996, again by The Institute for Genomic Research.[citation needed]
The development of new technologies has made genome sequencing dramatically cheaper and easier, and the number of complete genome sequences is growing rapidly. The US National Institutes of Health maintains one of several comprehensive databases of genomic information.[17] Among the thousands of completed genome sequencing projects include those for rice, a mouse, the plant Arabidopsis thaliana, the puffer fish, and the bacteria E. coli. In December 2013, scientists first sequenced the entire genome of a Neanderthal, an extinct species of humans. The genome was extracted from the toe bone of a 130,000-year-old Neanderthal found in a Siberian cave.[18][19]
New sequencing technologies, such as massive parallel sequencing have also opened up the prospect of personal genome sequencing as a diagnostic tool, as pioneered by Manteia Predictive Medicine. A major step toward that goal was the completion in 2007 of the full genome of James D. Watson, one of the co-discoverers of the structure of DNA.[20]
Whereas a genome sequence lists the order of every DNA base in a genome, a genome map identifies the landmarks. A genome map is less detailed than a genome sequence and aids in navigating around the genome. The Human Genome Project was organized to map and to sequence the human genome. A fundamental step in the project was the release of a detailed genomic map by Jean Weissenbach and his team at the Genoscope in Paris.[21][22]
Reference genome sequences and maps continue to be updated, removing errors and clarifying regions of high allelic complexity.[23] The decreasing cost of genomic mapping has permitted genealogical sites to offer it as a service,[24] to the extent that one may submit one's genome to crowdsourced scientific endeavours such as DNA.LAND at the New York Genome Center,[25] an example both of the economies of scale and of citizen science.[26]
Viral genomes
Viral genomes can be composed of either RNA or DNA. The genomes of RNA viruses can be either single-stranded RNA or double-stranded RNA, and may contain one or more separate RNA molecules (segments: monopartit or multipartit genome). DNA viruses can have either single-stranded or double-stranded genomes. Most DNA virus genomes are composed of a single, linear molecule of DNA, but some are made up of a circular DNA molecule.[27]
Prokaryotic genomes
Prokaryotes and eukaryotes have DNA genomes. Archaea and most bacteria have a single circular chromosome,[28] however, some bacterial species have linear or multiple chromosomes.[29][30] If the DNA is replicated faster than the bacterial cells divide, multiple copies of the chromosome can be present in a single cell, and if the cells divide faster than the DNA can be replicated, multiple replication of the chromosome is initiated before the division occurs, allowing daughter cells to inherit complete genomes and already partially replicated chromosomes. Most prokaryotes have very little repetitive DNA in their genomes.[31] However, some symbiotic bacteria (e.g. Serratia symbiotica) have reduced genomes and a high fraction of pseudogenes: only ~40% of their DNA encodes proteins.[32][33]
Some bacteria have auxiliary genetic material, also part of their genome, which is carried in plasmids. For this, the word genome should not be used as a synonym of chromosome.
Eukaryotic genomes

Eukaryotic genomes are composed of one or more linear DNA chromosomes. The number of chromosomes varies widely from Jack jumper ants and an asexual nemotode,[34] which each have only one pair, to a fern species that has 720 pairs.[35] It is surprising the amount of DNA that eukaryotic genomes contain compared to other genomes. The amount is even more than what is necessary for DNA protein-coding and noncoding genes due to the fact that eukaryotic genomes show as much as 64,000-fold variation in their sizes.[36] However, this special characteristic is caused by the presence of repetitive DNA, and transposable elements (TEs).
A typical human cell has two copies of each of 22 autosomes, one inherited from each parent, plus two sex chromosomes, making it diploid. Gametes, such as ova, sperm, spores, and pollen, are haploid, meaning they carry only one copy of each chromosome. In addition to the chromosomes in the nucleus, organelles such as the chloroplasts and mitochondria have their own DNA. Mitochondria are sometimes said to have their own genome often referred to as the "mitochondrial genome". The DNA found within the chloroplast may be referred to as the "plastome". Like the bacteria they originated from, mitochondria and chloroplasts have a circular chromosome.
Unlike prokaryotes where exon-intron organization of protein coding genes exists but is rather exceptional, eukaryotes generally have these features in their genes and their genomes contain variable amounts of repetitive DNA. In mammals and plants, the majority of the genome is composed of repetitive DNA.[37] Genes in eukaryotic genomes can be annotated using FINDER.[38] [39]
DNA sequencing
High-throughput technology makes sequencing to assemble new genomes accessible to everyone. Sequence polymorphisms are typically discovered by comparing resequenced isolates to a reference, whereas analyses of coverage depth and mapping topology can provide details regarding structural variations such as chromosomal translocations and segmental duplications.
Coding sequences
DNA sequences that carry the instructions to make proteins are referred to as coding sequences. The proportion of the genome occupied by coding sequences varies widely. A larger genome does not necessarily contain more genes, and the proportion of non-repetitive DNA decreases along with increasing genome size in complex eukaryotes.[37]

Noncoding sequences
Noncoding sequences include introns, sequences for non-coding RNAs, regulatory regions, and repetitive DNA. Noncoding sequences make up 98% of the human genome. There are two categories of repetitive DNA in the genome: tandem repeats and interspersed repeats.[40]
Tandem repeats
Short, non-coding sequences that are repeated head-to-tail are called tandem repeats. Microsatellites consisting of 2–5 basepair repeats, while minisatellite repeats are 30–35 bp. Tandem repeats make up about 4% of the human genome and 9% of the fruit fly genome.[41] Tandem repeats can be functional. For example, telomeres are composed of the tandem repeat TTAGGG in mammals, and they play an important role in protecting the ends of the chromosome.
In other cases, expansions in the number of tandem repeats in exons or introns can cause disease.[42] For example, the human gene huntingtin (Htt) typically contains 6–29 tandem repeats of the nucleotides CAG (encoding a polyglutamine tract). An expansion to over 36 repeats results in Huntington's disease, a neurodegenerative disease. Twenty human disorders are known to result from similar tandem repeat expansions in various genes. The mechanism by which proteins with expanded polygulatamine tracts cause death of neurons is not fully understood. One possibility is that the proteins fail to fold properly and avoid degradation, instead accumulating in aggregates that also sequester important transcription factors, thereby altering gene expression.[42]
Tandem repeats are usually caused by slippage during replication, unequal crossing-over and gene conversion.[43]
Transposable elements
Transposable elements (TEs) are sequences of DNA with a defined structure that are able to change their location in the genome.[41][31][44] TEs are categorized as either as a mechanism that replicates by copy-and-paste or as a mechanism that can be excised from the genome and inserted at a new location. In the human genome, there are three important classes of TEs that make up more than 45% of the human DNA; these classes are The long interspersed nuclear elements (LINEs), The interspersed nuclear elements (SINEs), and endogenous retroviruses. These elements have a big potential to modify the genetic control in a host organism.[36]
The movement of TEs is a driving force of genome evolution in eukaryotes because their insertion can disrupt gene functions, homologous recombination between TEs can produce duplications, and TE can shuffle exons and regulatory sequences to new locations.[45]
Retrotransposons
Retrotransposons[46] are found mostly in eukaryotes but not found in prokaryotes. Retrotransposons form a large portion of the genomes of many eukaryotes. A retrotransposon is a transposable element that transposes through an RNA intermediate. Retrotransposons[47] are composed of DNA, but are transcribed into RNA for transposition, then the RNA transcript is copied back to DNA formation with the help of a specific enzyme called reverse transcriptase. A retrotransposon that carries reverse transcriptase in its sequence can trigger its own transposition but retrotransposons that lack a reverse transcriptase must use reverse transcriptase synthesized by another retrotransposon. Retrotransposons can be transcribed into RNA, which are then duplicated at another site into the genome.[48] Retrotransposons can be divided into long terminal repeats (LTRs) and non-long terminal repeats (Non-LTRs).[45]
Long terminal repeats (LTRs) are derived from ancient retroviral infections, so they encode proteins related to retroviral proteins including gag (structural proteins of the virus), pol (reverse transcriptase and integrase), pro (protease), and in some cases env (envelope) genes.[44] These genes are flanked by long repeats at both 5' and 3' ends. It has been reported that LTRs consist of the largest fraction in most plant genome and might account for the huge variation in genome size.[49]
Non-long terminal repeats (Non-LTRs) are classified as long interspersed nuclear elements (LINEs), short interspersed nuclear elements (SINEs), and Penelope-like elements (PLEs). In Dictyostelium discoideum, there is another DIRS-like elements belong to Non-LTRs. Non-LTRs are widely spread in eukaryotic genomes.[50]
Long interspersed elements (LINEs) encode genes for reverse transcriptase and endonuclease, making them autonomous transposable elements. The human genome has around 500,000 LINEs, taking around 17% of the genome.[51]
Short interspersed elements (SINEs) are usually less than 500 base pairs and are non-autonomous, so they rely on the proteins encoded by LINEs for transposition.[52] The Alu element is the most common SINE found in primates. It is about 350 base pairs and occupies about 11% of the human genome with around 1,500,000 copies.[45]
DNA transposons
DNA transposons encode a transposase enzyme between inverted terminal repeats. When expressed, the transposase recognizes the terminal inverted repeats that flank the transposon and catalyzes its excision and reinsertion in a new site.[41] This cut-and-paste mechanism typically reinserts transposons near their original location (within 100 kb).[45] DNA transposons are found in bacteria and make up 3% of the human genome and 12% of the genome of the roundworm C. elegans.[45]
Genome size
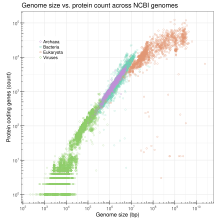
Genome size is the total number of the DNA base pairs in one copy of a haploid genome. Genome size varies widely across species. Invertebrates have small genomes, this is also correlated to a small number of transposable elements. Fish and Amphibians have intermediate-size genomes, and birds have relatively small genomes but it has been suggested that birds lost a substantial portion of their genomes during the phase of transition to flight. Before this loss, DNA methylation allows the adequate expansion of the genome.[36]
In humans, the nuclear genome comprises approximately 3.1 billion nucleotides of DNA, divided into 24 linear molecules, the shortest 45 000 000 nucleotides in length and the longest 248 000 000 nucleotides, each contained in a different chromosome.[53] There is no clear and consistent correlation between morphological complexity and genome size in either prokaryotes or lower eukaryotes.[37][54] Genome size is largely a function of the expansion and contraction of repetitive DNA elements.
Since genomes are very complex, one research strategy is to reduce the number of genes in a genome to the bare minimum and still have the organism in question survive. There is experimental work being done on minimal genomes for single cell organisms as well as minimal genomes for multi-cellular organisms (see developmental biology). The work is both in vivo and in silico.[55][56]
Genome size differences due to transposable elements

There are many enormous differences in size in genomes, specially mentioned before in the multicellular eukaryotic genomes. Much of this is due to the differing abundances of transposable elements, which evolve by creating new copies of themselves in the chromosomes.[36] Eukaryote genomes often contain many thousands of copies of these elements, most of which have acquired mutations that make them defective. Here is a table of some significant or representative genomes. See #See also for lists of sequenced genomes.
| Organism type | Organism | Genome size (base pairs) |
Approx. no. of genes | Note | |
|---|---|---|---|---|---|
| Virus | Porcine circovirus type 1 | 1,759 | 1.8 kB | Smallest viruses replicating autonomously in eukaryotic cells[57] | |
| Virus | Bacteriophage MS2 | 3,569 | 3.6 kB | First sequenced RNA-genome[58] | |
| Virus | SV40 | 5,224 | 5.2 kB | [59] | |
| Virus | Phage Φ-X174 | 5,386 | 5.4 kB | First sequenced DNA-genome[60] | |
| Virus | HIV | 9,749 | 9.7 kB | [61] | |
| Virus | Phage λ | 48,502 | 48.5 kB | Often used as a vector for the cloning of recombinant DNA | |
| Virus | Megavirus | 1,259,197 | 1.3 MB | Until 2013 the largest known viral genome[65] | |
| Virus | Pandoravirus salinus | 2,470,000 | 2.47 MB | Largest known viral genome.[66] | |
| Eukaryotic organelle | Human mitochondrion | 16,569 | 16.6 kB | [67] | |
| Bacterium | Nasuia deltocephalinicola (strain NAS-ALF) | 112,091 | 112 kB | 137 | Smallest known non-viral genome. Symbiont of leafhoppers.[68] |
| Bacterium | Carsonella ruddii | 159,662 | 160 kB | An endosymbiont of psyllid insects | |
| Bacterium | Buchnera aphidicola | 600,000 | 600 kB | An endosymbiont of aphids[69] | |
| Bacterium | Wigglesworthia glossinidia | 700,000 | 700 kB | A symbiont in the gut of the tsetse fly | |
| Bacterium – cyanobacterium | Prochlorococcus spp. (1.7 Mb) | 1,700,000 | 1.7 MB | 1,884 | Smallest known cyanobacterium genome. One of the primary photosynthesizers on Earth.[70][71] |
| Bacterium | Haemophilus influenzae | 1,830,000 | 1.8 MB | First genome of a living organism sequenced, July 1995[72] | |
| Bacterium | Escherichia coli | 4,600,000 | 4.6 MB | 4,288 | [73] |
| Bacterium – cyanobacterium | Nostoc punctiforme | 9,000,000 | 9 MB | 7,432 | 7432 open reading frames[74] |
| Bacterium | Solibacter usitatus (strain Ellin 6076) | 9,970,000 | 10 MB | [75] | |
| Amoeboid | Polychaos dubium ("Amoeba" dubia) | 670,000,000,000 | 670 GB | Largest known genome.[76] (Disputed)[77] | |
| Plant | Genlisea tuberosa | 61,000,000 | 61 MB | Smallest recorded flowering plant genome, 2014[78] | |
| Plant | Arabidopsis thaliana | 135,000,000[79] | 135 MB | 27,655[80] | First plant genome sequenced, December 2000[81] |
| Plant | Populus trichocarpa | 480,000,000 | 480 MB | 73,013 | First tree genome sequenced, September 2006[82] |
| Plant | Pinus taeda (Loblolly pine) | 22,180,000,000 | 22.18 GB | 50,172 | Gymnosperms generally have much larger genomes than angiosperms[83][84] |
| Plant | Fritillaria assyriaca | 130,000,000,000 | 130 GB | ||
| Plant | Paris japonica (Japanese-native, order Liliales) | 150,000,000,000 | 150 GB | Largest plant genome known[85] | |
| Plant – moss | Physcomitrella patens | 480,000,000 | 480 MB | First genome of a bryophyte sequenced, January 2008[86] | |
| Fungus – yeast | Saccharomyces cerevisiae | 12,100,000 | 12.1 MB | 6,294 | First eukaryotic genome sequenced, 1996[87] |
| Fungus | Aspergillus nidulans | 30,000,000 | 30 MB | 9,541 | [88] |
| Nematode | Pratylenchus coffeae | 20,000,000 | 20 MB | [89] Smallest animal genome known[90] | |
| Nematode | Caenorhabditis elegans | 100,300,000 | 100 MB | 19,000 | First multicellular animal genome sequenced, December 1998[91] |
| Insect | Belgica antarctica (Antarctic midge) | 99,000,000 | 99 MB | Smallest insect genome sequenced thus far, likely an adaptation to an extreme environment[92] | |
| Insect | Drosophila melanogaster (fruit fly) | 175,000,000 | 175 MB | 13,600 | Size variation based on strain (175–180 Mb; standard y w strain is 175 Mb)[93] |
| Insect | Apis mellifera (honey bee) | 236,000,000 | 236 MB | 10,157 | [94] |
| Insect | Bombyx mori (silk moth) | 432,000,000 | 432 MB | 14,623 | 14,623 predicted genes[95] |
| Insect | Solenopsis invicta (fire ant) | 480,000,000 | 480 MB | 16,569 | [96] |
| Crustacean | Zdroj:https://en.wikipedia.org?pojem=Genome|||||
